| Map > Problem Definition > Data Preparation > Data Exploration > Bivariate |
Data Exploration - Bivariate Analysis |
| I. Data Preparation |
|
1- Load libraries |
|
library(data.table) library(formattable) library(plotrix) library(limma) |
|
2- Read Expressions file |
|
df <- read.csv("GSE74763_rawlog_expr.csv") df2 <- df[,-1] rownames(df2) <- df[,1] expr <- transpose(df2) rownames(expr) <- colnames(df2) colnames(expr) <- rownames(df2) dim(expr) |
|
3- Read Samples file |
|
targets <- read.csv("GSE74763_rawlog_targets.csv") colnames(targets) dim(targets) |
|
4- Merge Expressions with Samples |
|
data <- cbind(expr, targets) colnames(data) dim(data) |
| II. Numerical & Numerical |
| 1- Linear Correlation |
| cor(data$P000001,data$P000002) |
|
|
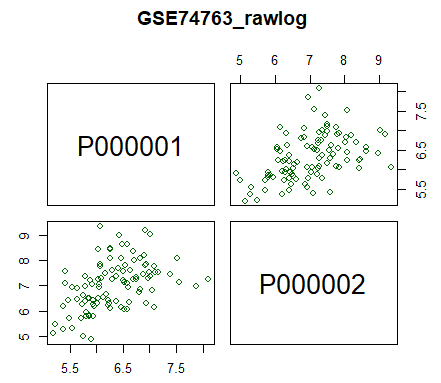
| 2- Scatter plot |
| pairs(~P000001+P000002, data=data, main="GSE74763_rawlog", col="darkgreen") |
|
|
| III. Categorical & Categorical |
| 1- Count (crosstab) |
| xtabs(~target+gender, data=data) |
|
|
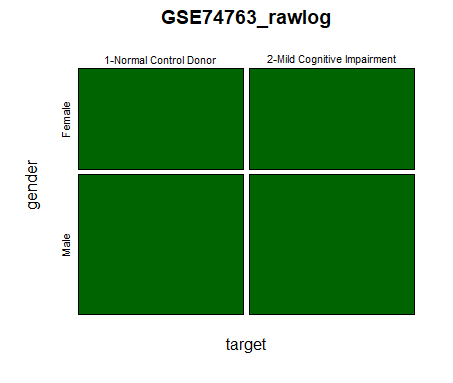
| 2- CrossTab Plot |
| xt <-
xtabs(~target+gender, data=data) plot(xt, main="GSE74763_rawlog", col="darkgreen") |
|
|
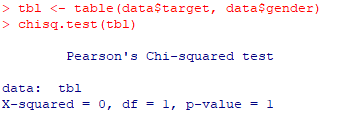
| 3- Chi2 test |
| tbl <-
table(data$target, data$gender) chisq.test(tbl) |
|
|
| IV. Categorical & Numerical |
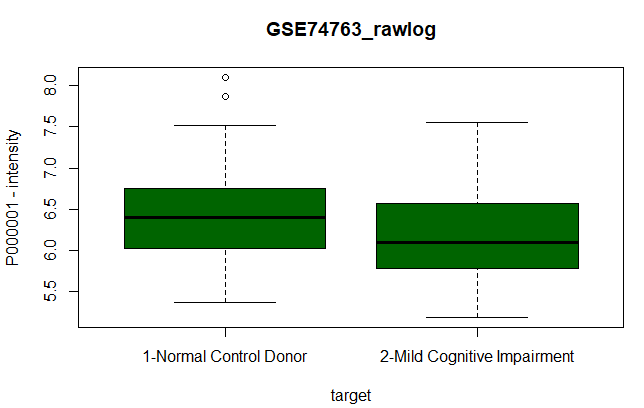
| 1- Box plot |
| boxplot(P000001~target, data=data, col="darkgreen", main="GSE74763_rawlog", ylab="P000001 - intensity") |
|
|
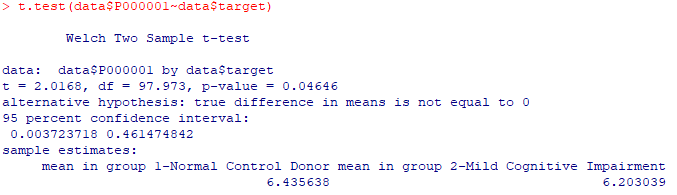
| 2- t-test |
| t.test(data$P000002~data$target) |
|
|
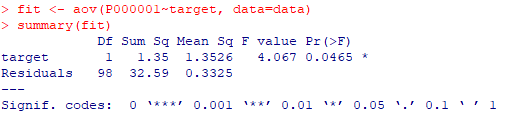
| 3- ANOVA |
| fit <-
aov(P000001~target, data=data) summary(fit) |
|
|
| V. Bioada SmartArray |
| Watch this video to learn how you can perform bivariate analysis using Bioada SmartArray significantly faster and easier. |