| Map > Problem Definition > Data Preparation > Data Exploration > Univariate |
Data Exploration - Univariate Analysis |
| I. Data Preparation |
|
1- Load libraries |
|
library(data.table) library(formattable) library(plotrix) library(limma) |
|
2- Read Expressions file |
|
df <- read.csv("GSE74763_rawlog_expr.csv") df2 <- df[,-1] rownames(df2) <- df[,1] expr <- transpose(df2) rownames(expr) <- colnames(df2) colnames(expr) <- rownames(df2) dim(expr) |
|
3- Read Samples file |
|
targets <- read.csv("GSE74763_rawlog_targets.csv") colnames(targets) dim(targets) |
|
4- Merge Expressions with Samples |
|
data <- cbind(expr, targets) colnames(data) dim(data) |
| II. Categorical Variables |
| 1- Count and Count% |
|
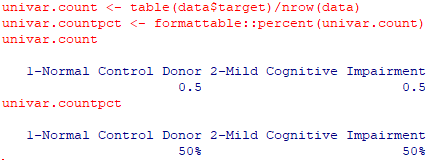
univar.count <- table(data$target)/nrow(data) univar.countpct <- formattable::percent(univar.count) univar.count univar.countpct |
|
|
| 2- Pie and Bar charts |
|
pie(table(data$target), main =
"target") lbl <- unique(data$target) pie3D(table(data$target),labels=lbl, explode = 0.1, main = "target") barplot(table(data$target), main="target", xlab="", ylab="Count", col="blue") |
|
|
| III. Numerical Variables |
| 1- Summary (Descriptive Statistics) |
| summary(data$age) |
|
|
| 2- Plots |
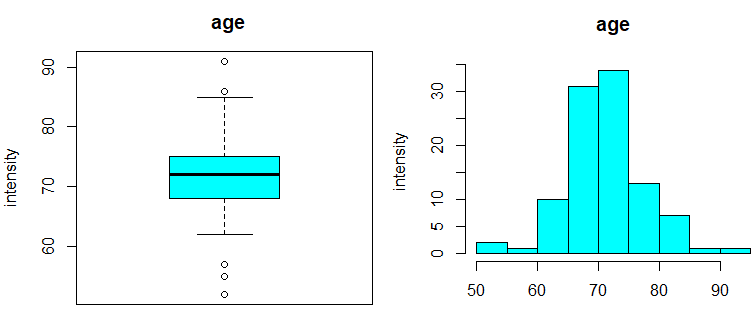
| boxplot(data$age, col="cyan",
main="age", ylab="intensity") hist(data$age, col="cyan", main="age", ylab="intensity") plotDensities(data$age, main="age", col="blue") |
|
|
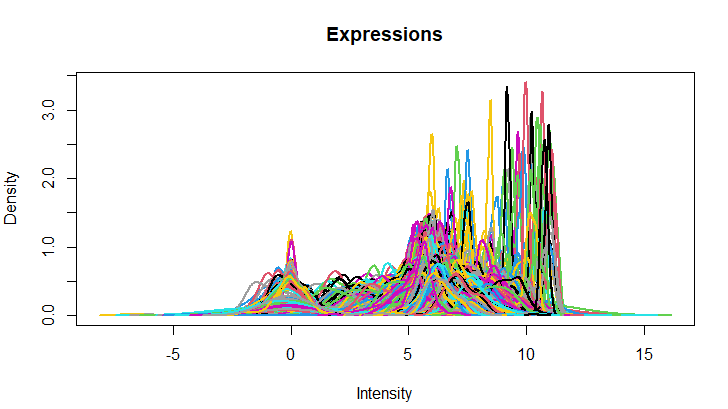
| 3- Density plot for all expressions |
| plotDensities(expr, main="Expressions", legend=F) |
|
|
| IV. Bioada SmartArray |
| This video shows how you can perform univariate analysis using Bioada SmartArray significantly faster and easier. |