Introduction Last updated: 2020-07-31
There are many factors that make data preparation challenging, from understanding where to find the data to extracting it, then properly formatting it and finally loading it to a database management system (DBMS). SmartArray reduces data preparation time, so that you the focus is on using a high quality and enriched data for visualization, statistical analysis and predictive modeling. With Bioada, users have the full control on the data.
SmartArray provides a unique interactive data exploration platform for ad-hoc queries, data visualizations and statistical analysis. Users can explore data interactively and apply statistical models and analytical techniques to find potential biomarkers in the genomic data.Users can focus and emphasize on interactivity and effective integration of techniques from data science.
Genomic datasets are high-dimensional molecular profiles pose challenges to data interpretation and hypothesis generation. SmartArray has a method that discovers significantly enriched pathways and gene ontology terms across datasets. SmartArray enrichment is a versatile method that improves systems-level understanding of cellular entities in health and disease through integration of genomic datasets and pathway and gene ontology annotations

 "User" icon.A message box appears as below.
"User" icon.A message box appears as below. button.All the Inactive users will be displayed under "Inactive Users" list .
button.All the Inactive users will be displayed under "Inactive Users" list .
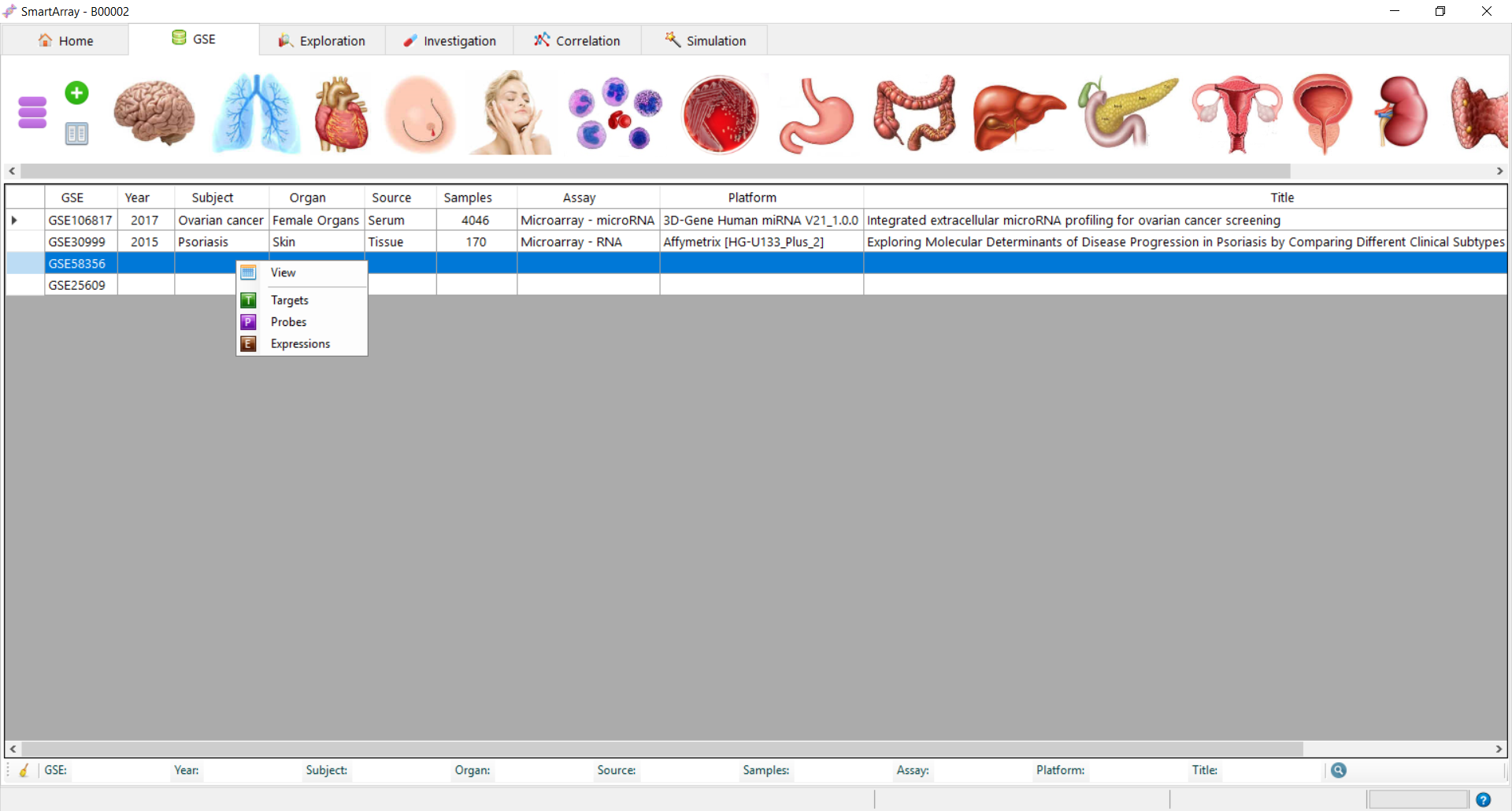
 button.It loads the existing databases in the local database server.
button.It loads the existing databases in the local database server.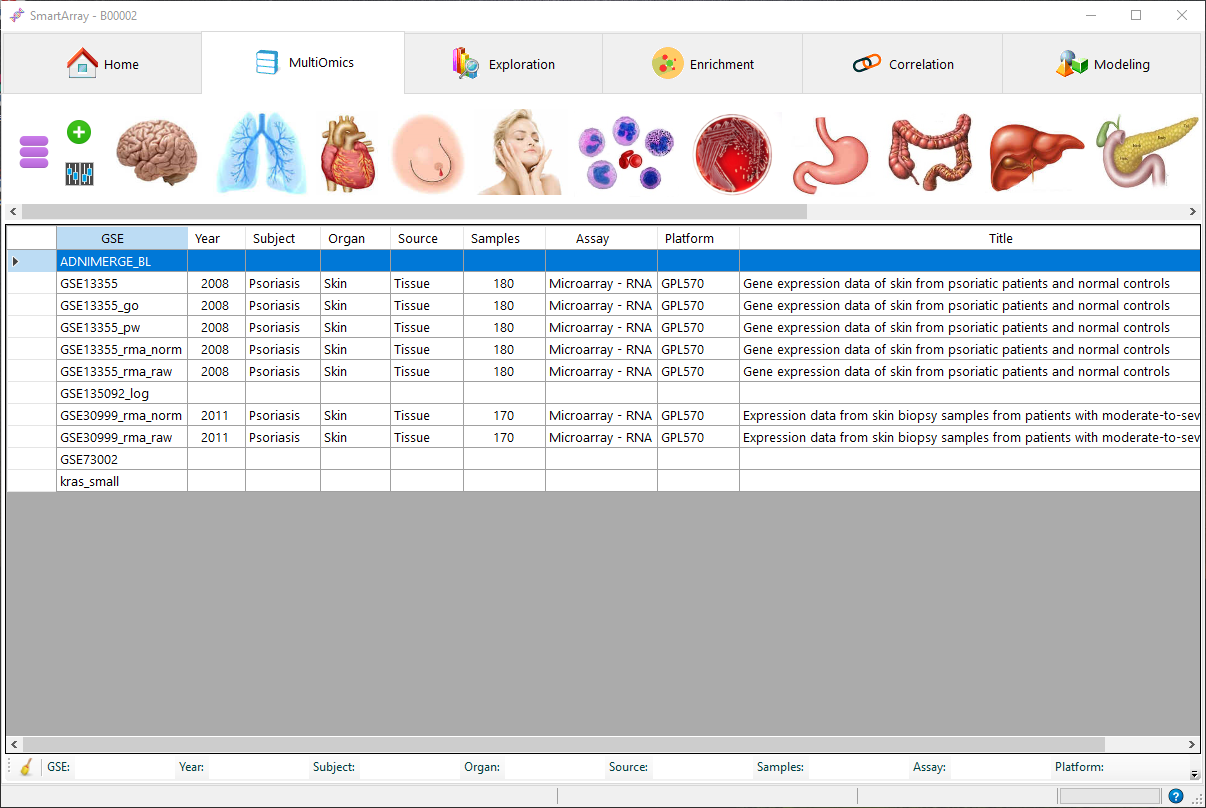
 to run the query and the results are as shown above.
to run the query and the results are as shown above. to save the query results.
to save the query results.  to get the aggreagte of the selected column.
to get the aggreagte of the selected column.  button.A new window is opened as shown below.
button.A new window is opened as shown below. . Enter new database name and click OK.
. Enter new database name and click OK. on to connect
on to connect  to upload the data to database.Click on
to upload the data to database.Click on  to view the file.
to view the file. 
 for Log transformation. This transforms the expression file to log expression and saves the file.Below is sample of log expression file.
for Log transformation. This transforms the expression file to log expression and saves the file.Below is sample of log expression file.  to replace any missing value with the average value of corresponding row. This modifies the expression file and saves the file.Below is sample of replaced file.
to replace any missing value with the average value of corresponding row. This modifies the expression file and saves the file.Below is sample of replaced file.  to delete any empty row . This changes the expression file and saves the file.Below is sample of clean file.
to delete any empty row . This changes the expression file and saves the file.Below is sample of clean file.  .A new compare window opens up.
.A new compare window opens up.
 to get the distribution of data from both datasets
to get the distribution of data from both datasets 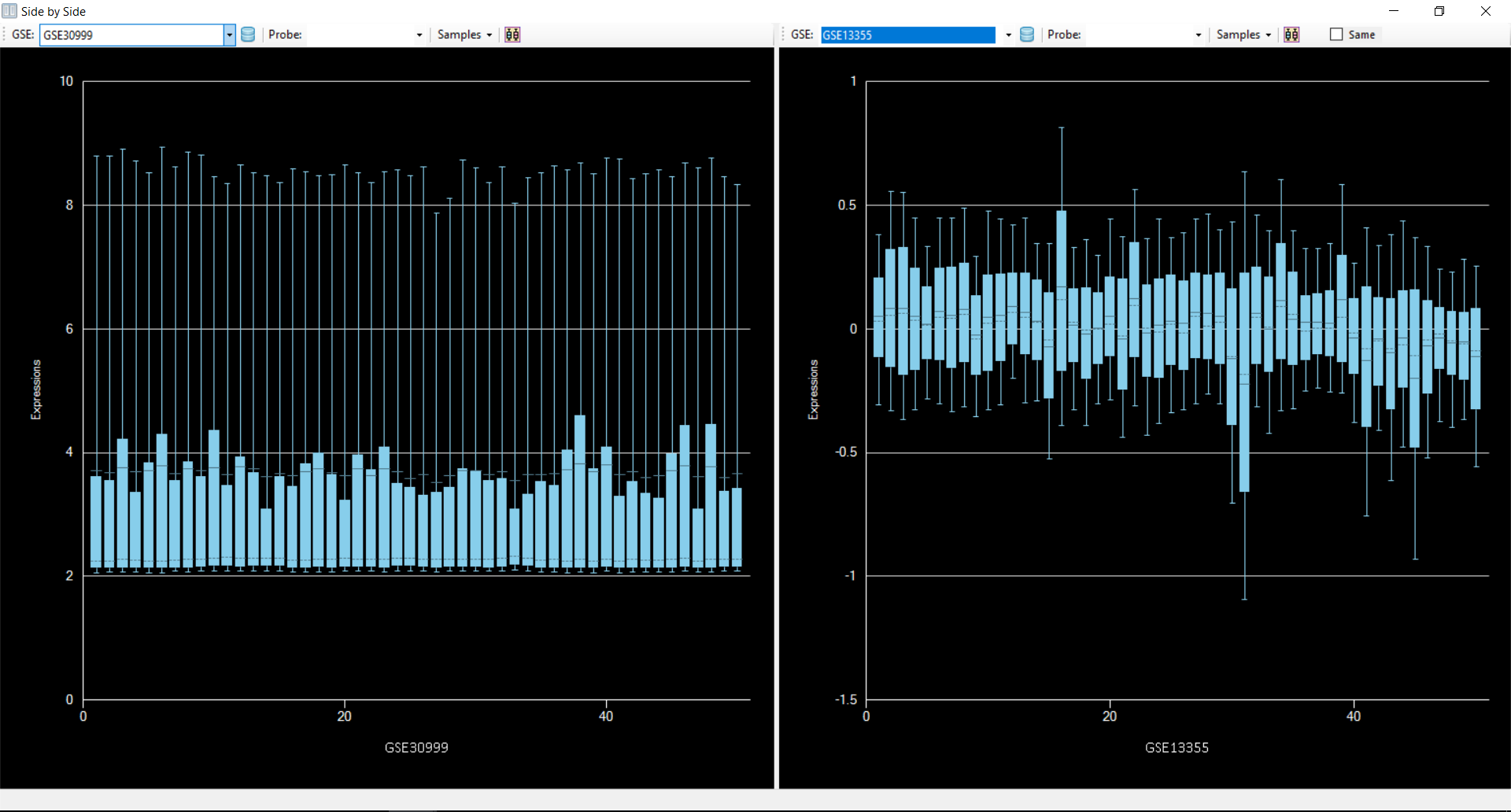
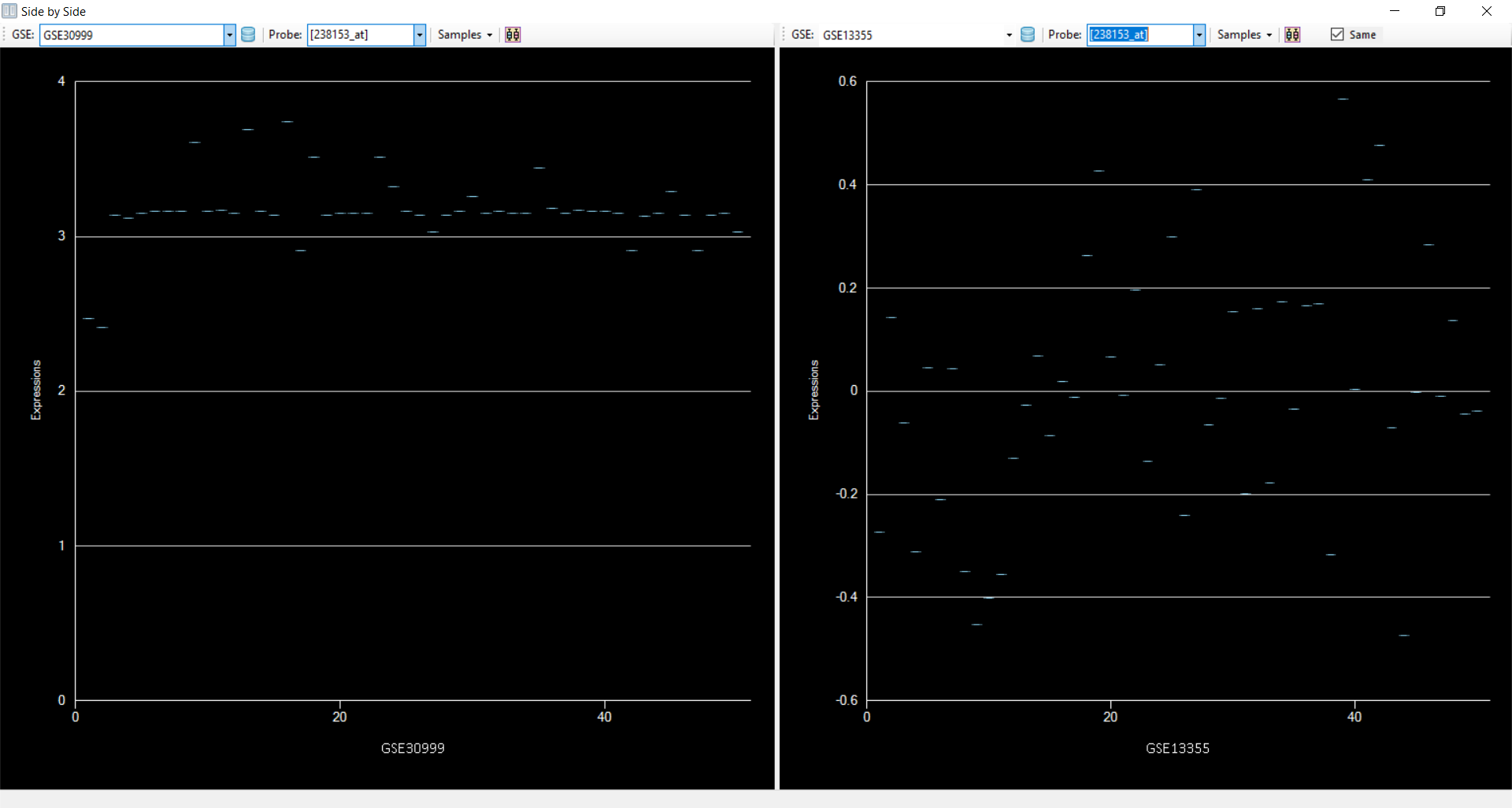
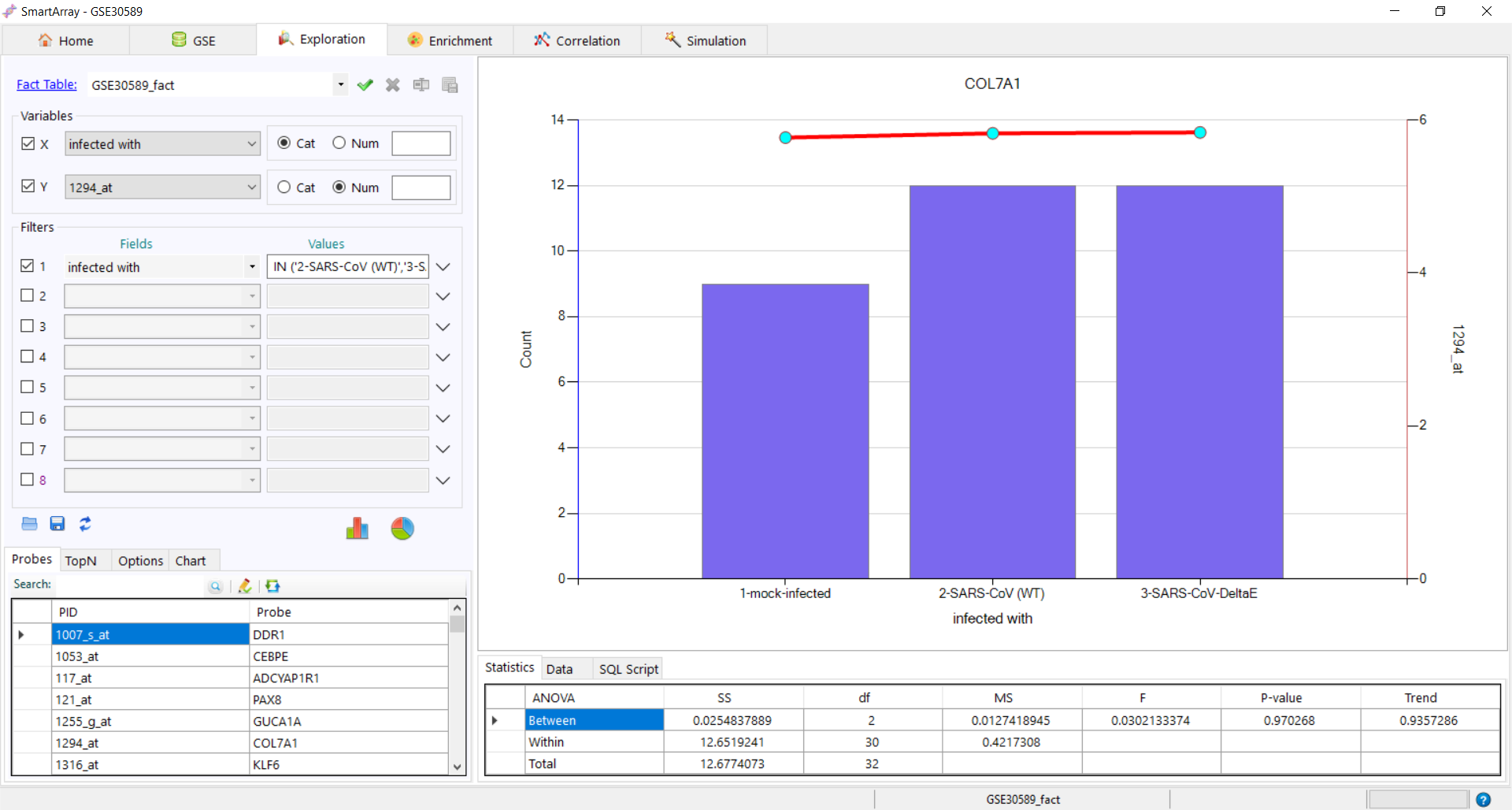
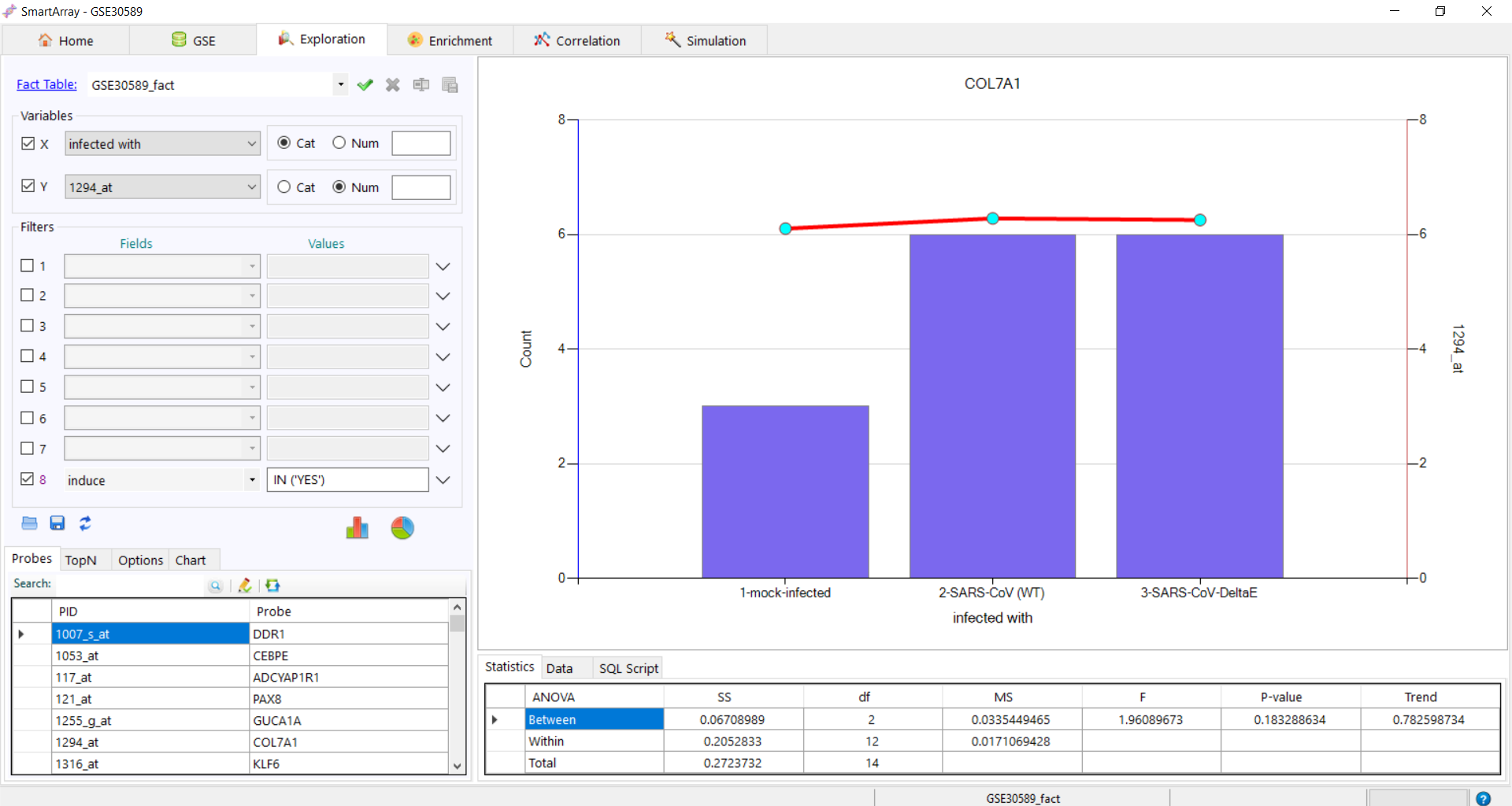
 Add Filter
Add Filter Refresh link to clear all the filters or click on "OK" to apply the filter.
Refresh link to clear all the filters or click on "OK" to apply the filter.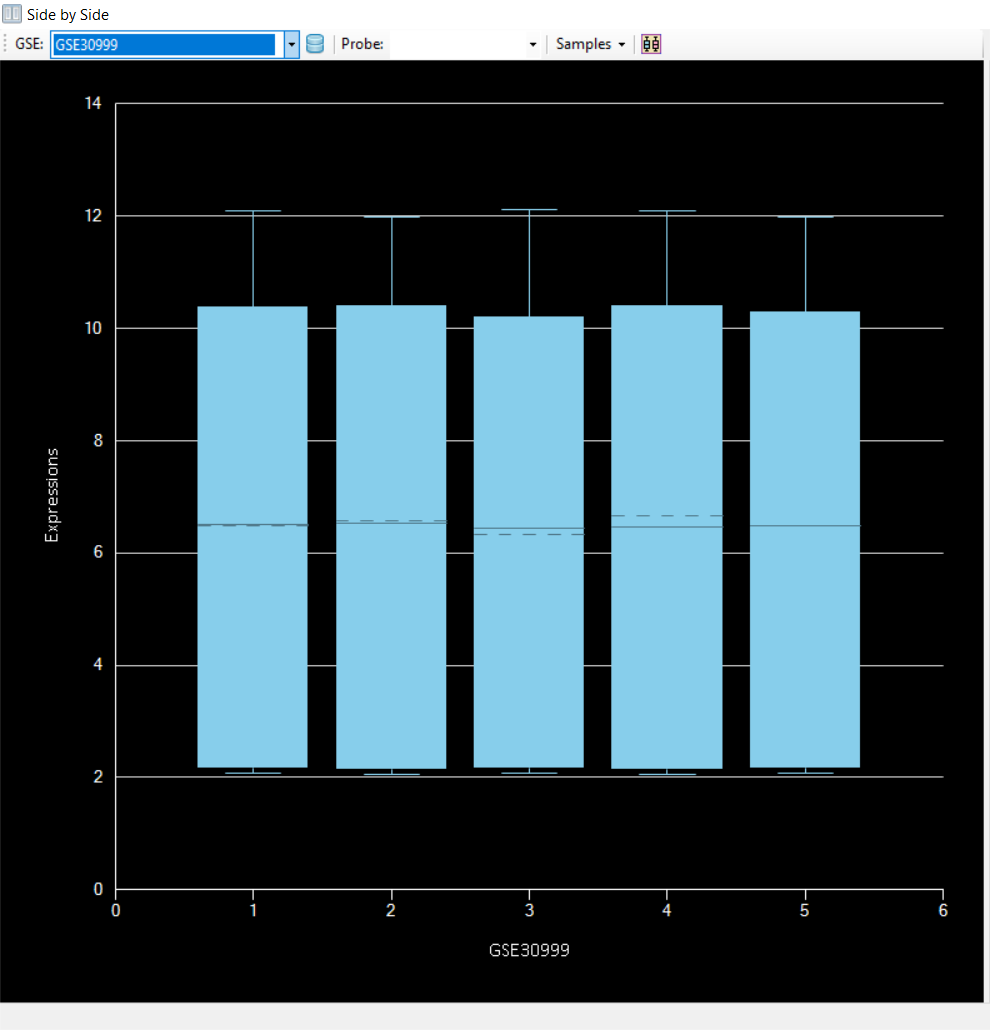
 View Filter to check the filters applied.
View Filter to check the filters applied. Delete Filter to delete the existing filter.
Delete Filter to delete the existing filter. 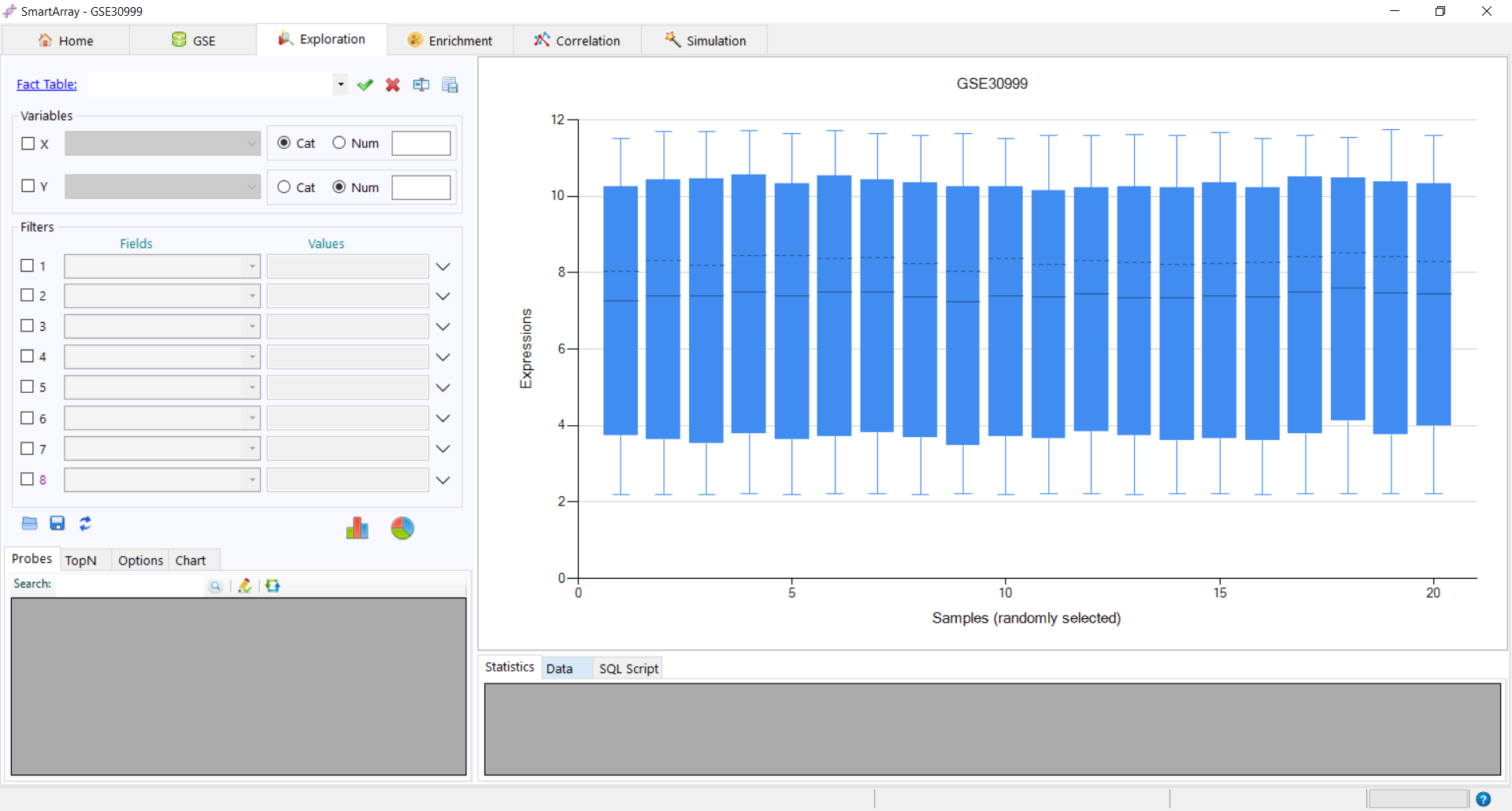
 to load the table. The original fact table cannot be deleted,renamed or exported.
to load the table. The original fact table cannot be deleted,renamed or exported. to delete the fact table.
to delete the fact table. to rename the fact table.
to rename the fact table. to export the fact table.
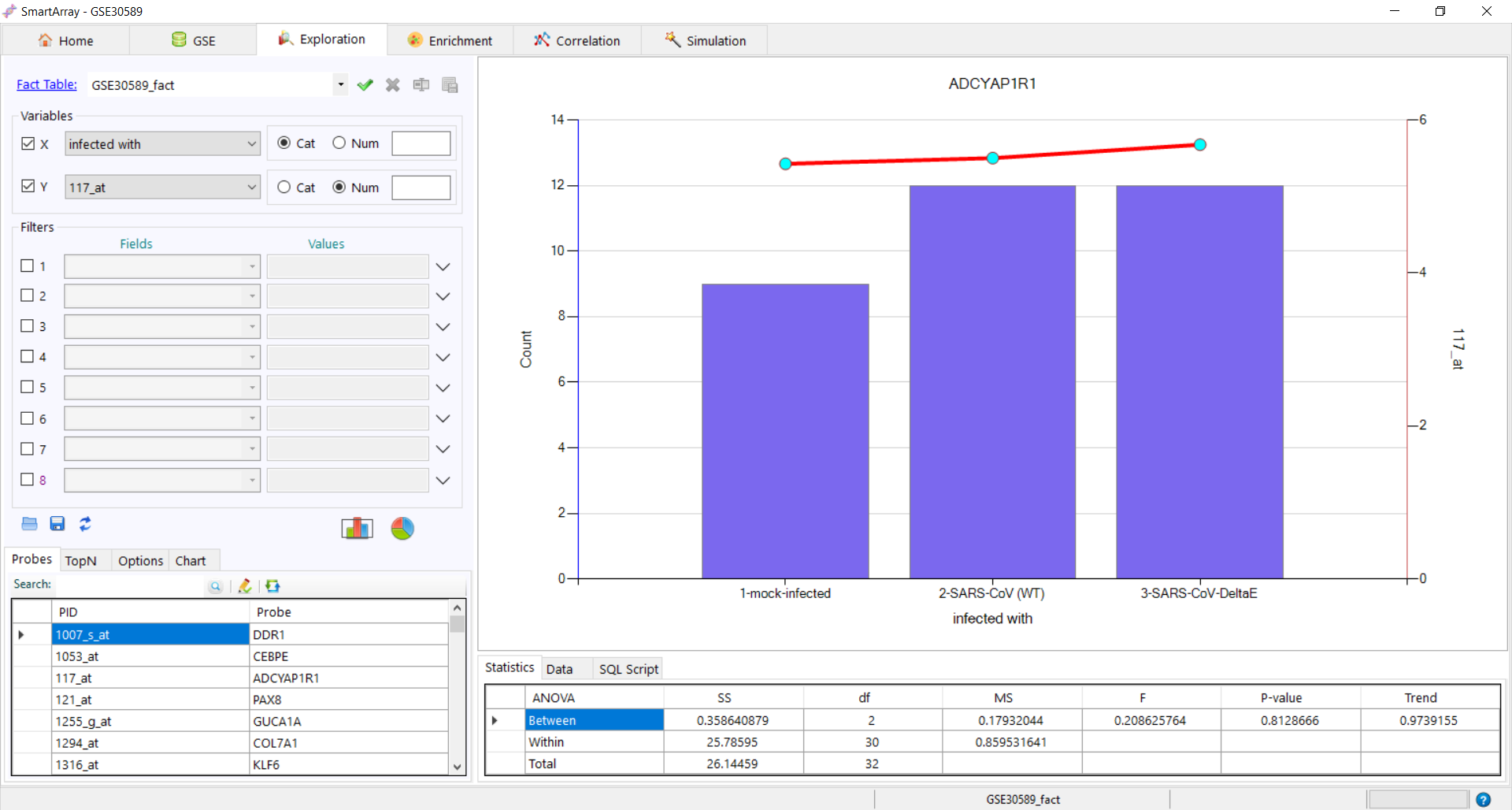
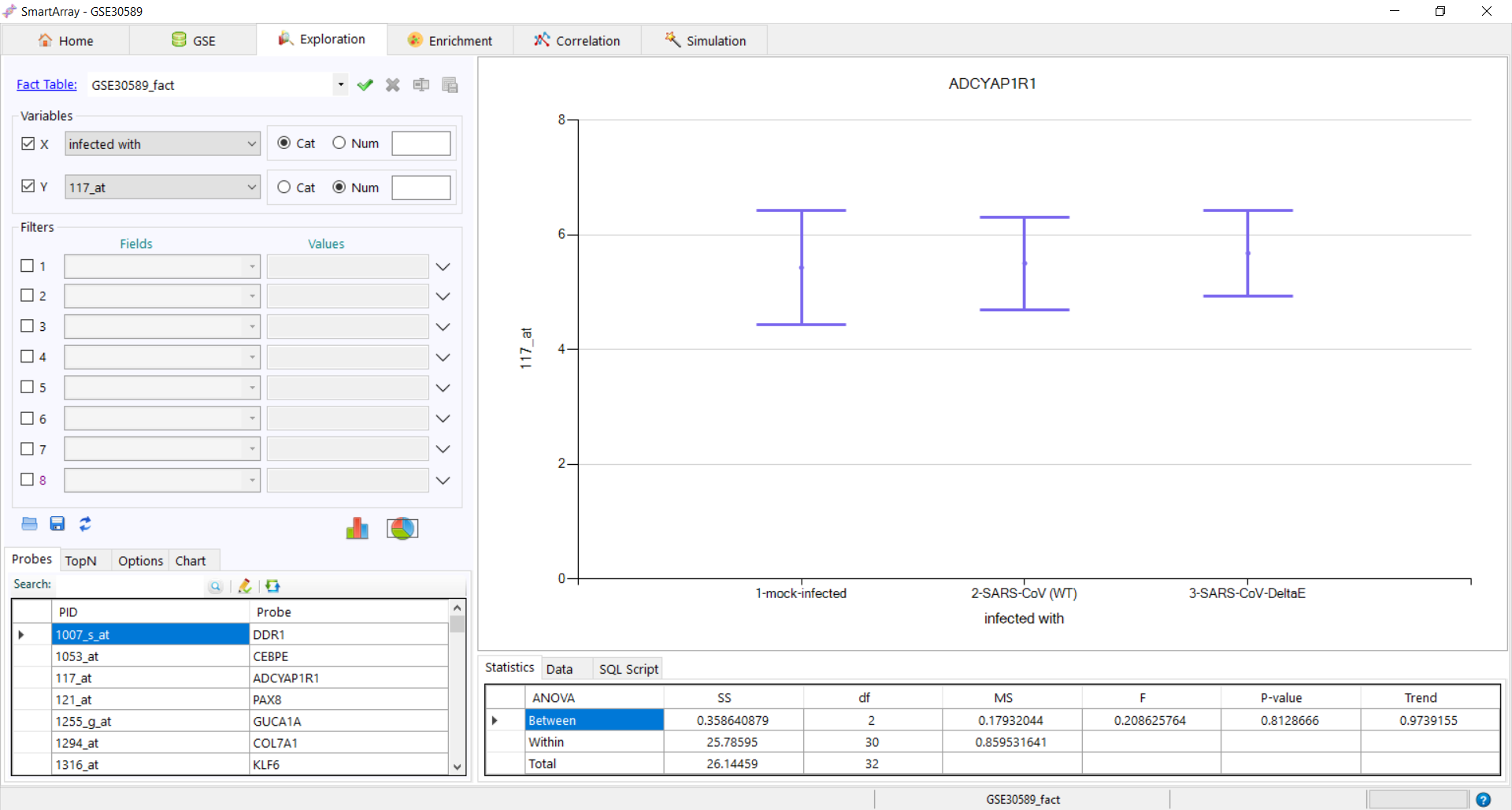
to export the fact table. to get the distribution of the variant. The data and the SQL Script used can be
seen at the bottom of the screen.
to get the distribution of the variant. The data and the SQL Script used can be
seen at the bottom of the screen.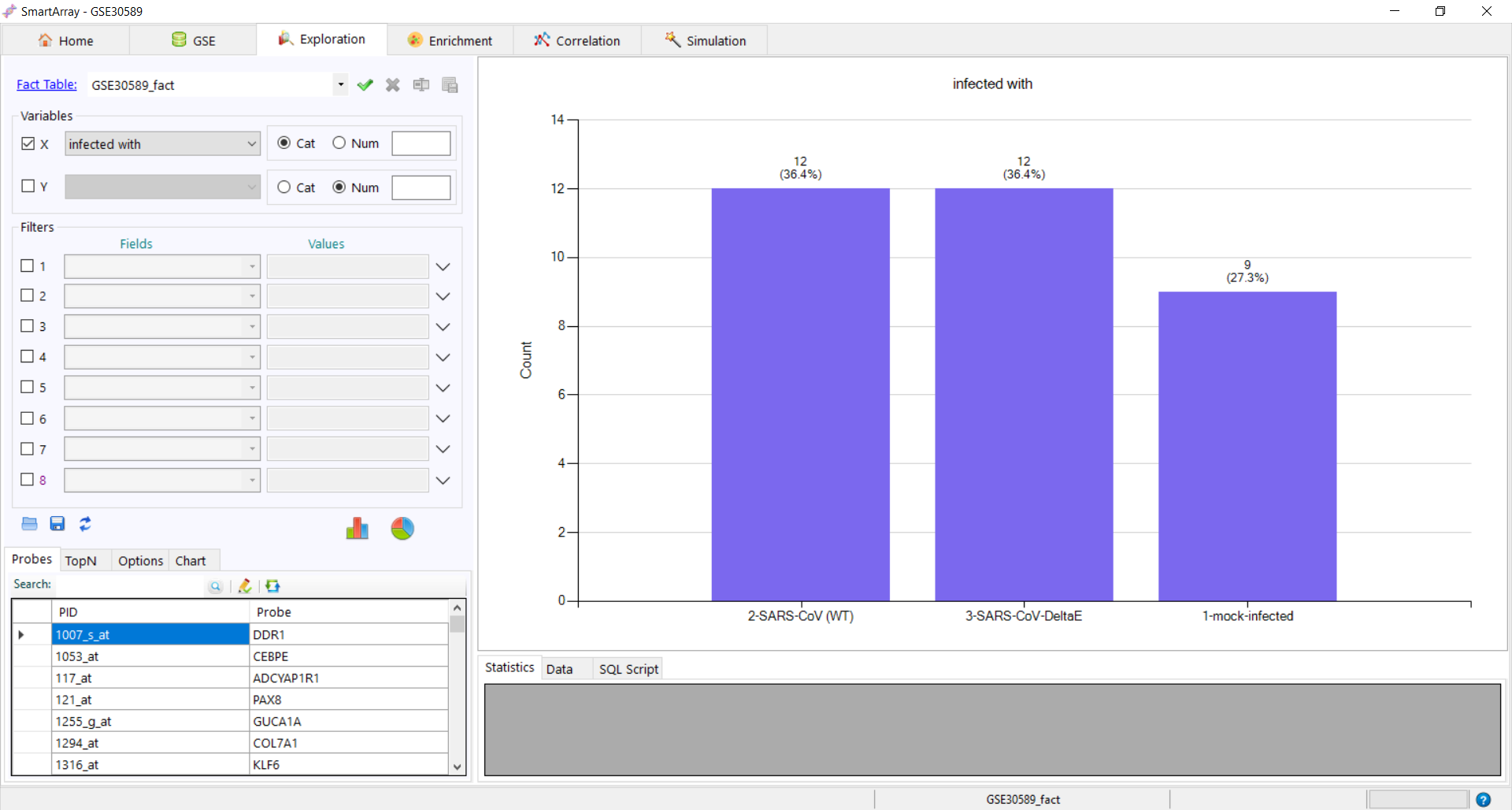
 to get the pie chart distribution.
to get the pie chart distribution. 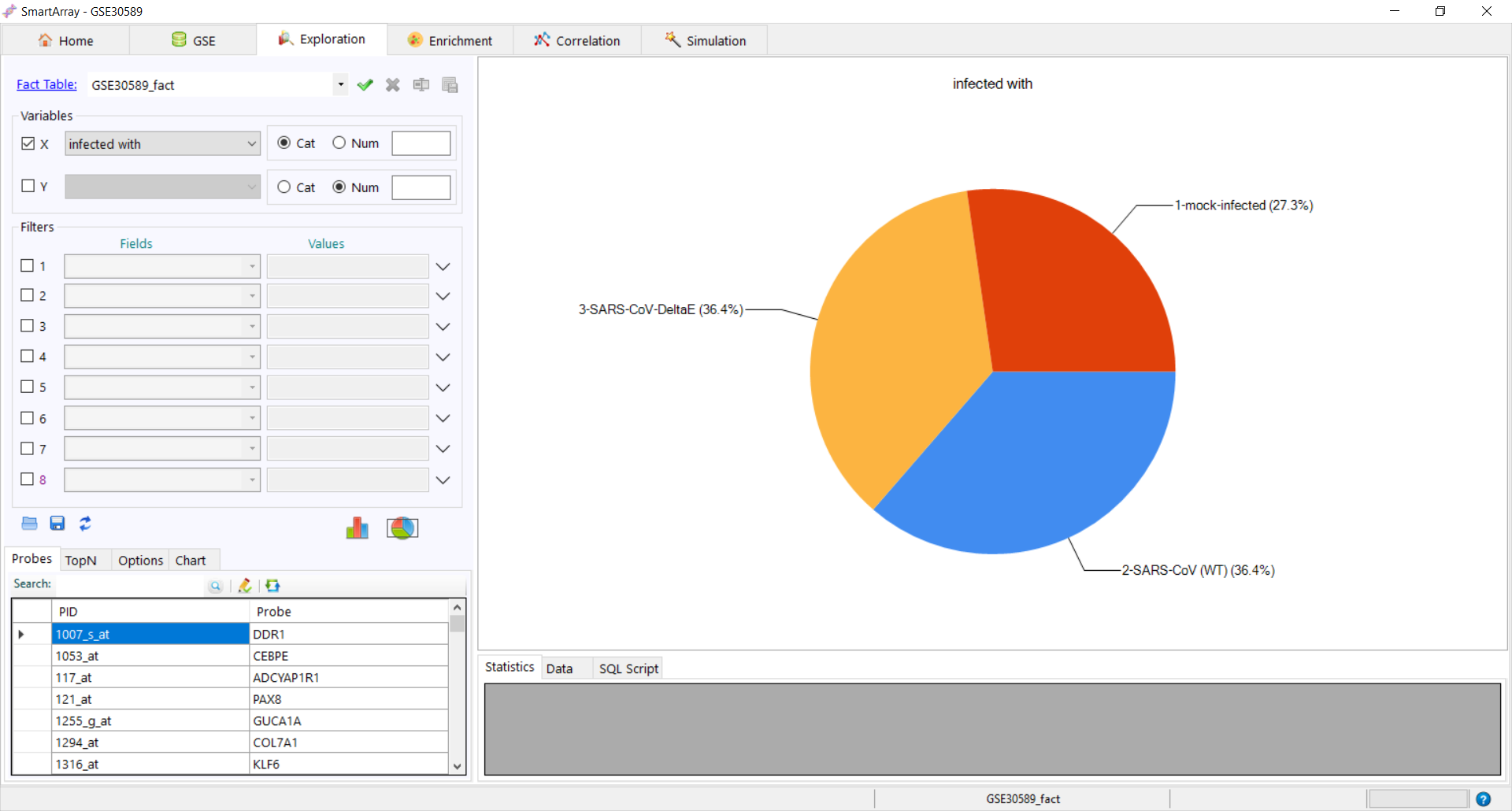
 to refresh the existing filters.
to refresh the existing filters.  to save the data.
to save the data.  to load the saved filters.
to load the saved filters.  to search the next matching value.
to search the next matching value. to rename the probe.
to rename the probe. to get the Volcano/Manhattan plot.
to get the Volcano/Manhattan plot.
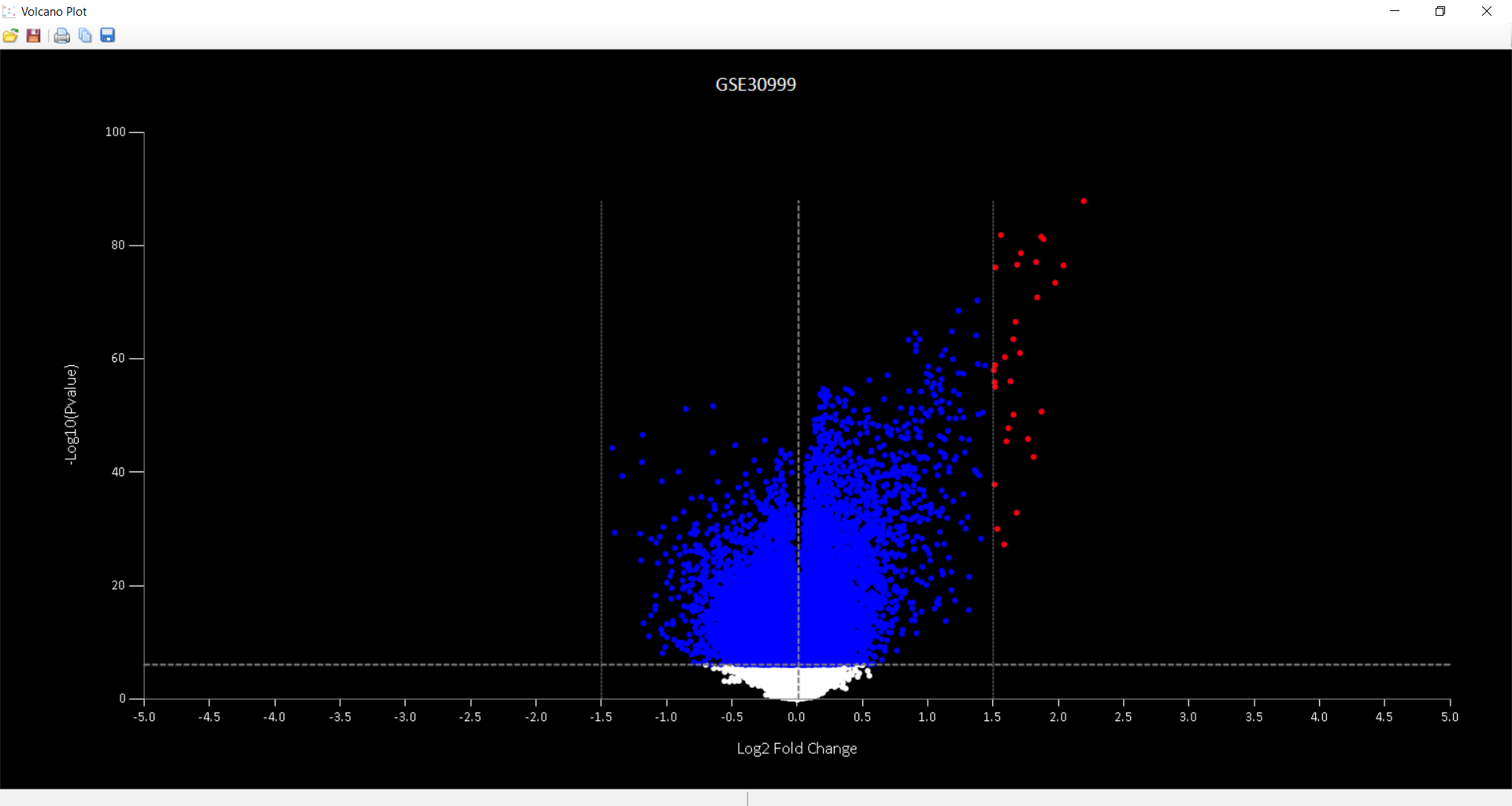
 to load the data used for obtaining volcano plot.
to load the data used for obtaining volcano plot.  to save the data used for obtaining volcano plot.
to save the data used for obtaining volcano plot.  to print the volcano plot.
to print the volcano plot.  to copy the volcano plot.
to copy the volcano plot. 
 to save the Top N data to database.The textbox can be used to name the table and the number of rows(N) to be saved. The saved dataset appears on the Fact_Table list
to save the Top N data to database.The textbox can be used to name the table and the number of rows(N) to be saved. The saved dataset appears on the Fact_Table list
 to show the saved plot
to show the saved plot 
 Overlap to see the probes.
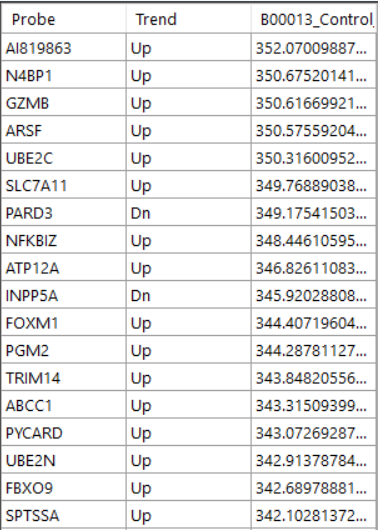
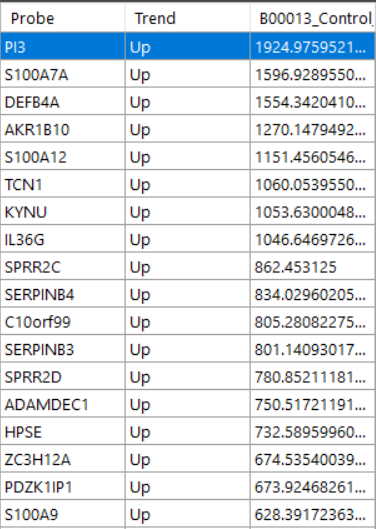
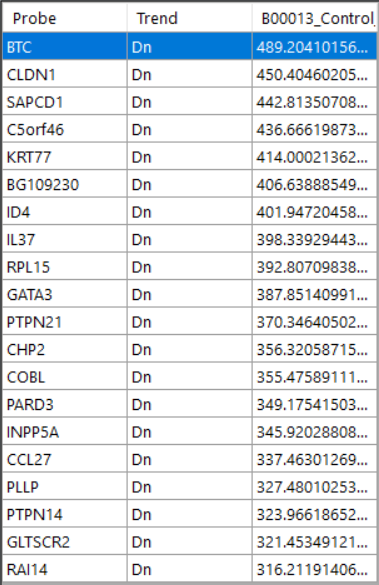
Overlap to see the probes. Heatmap to see the heatmap of all probes
Heatmap to see the heatmap of all probes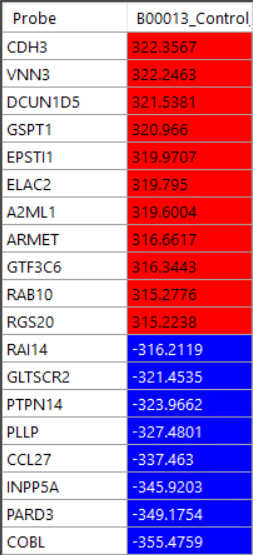
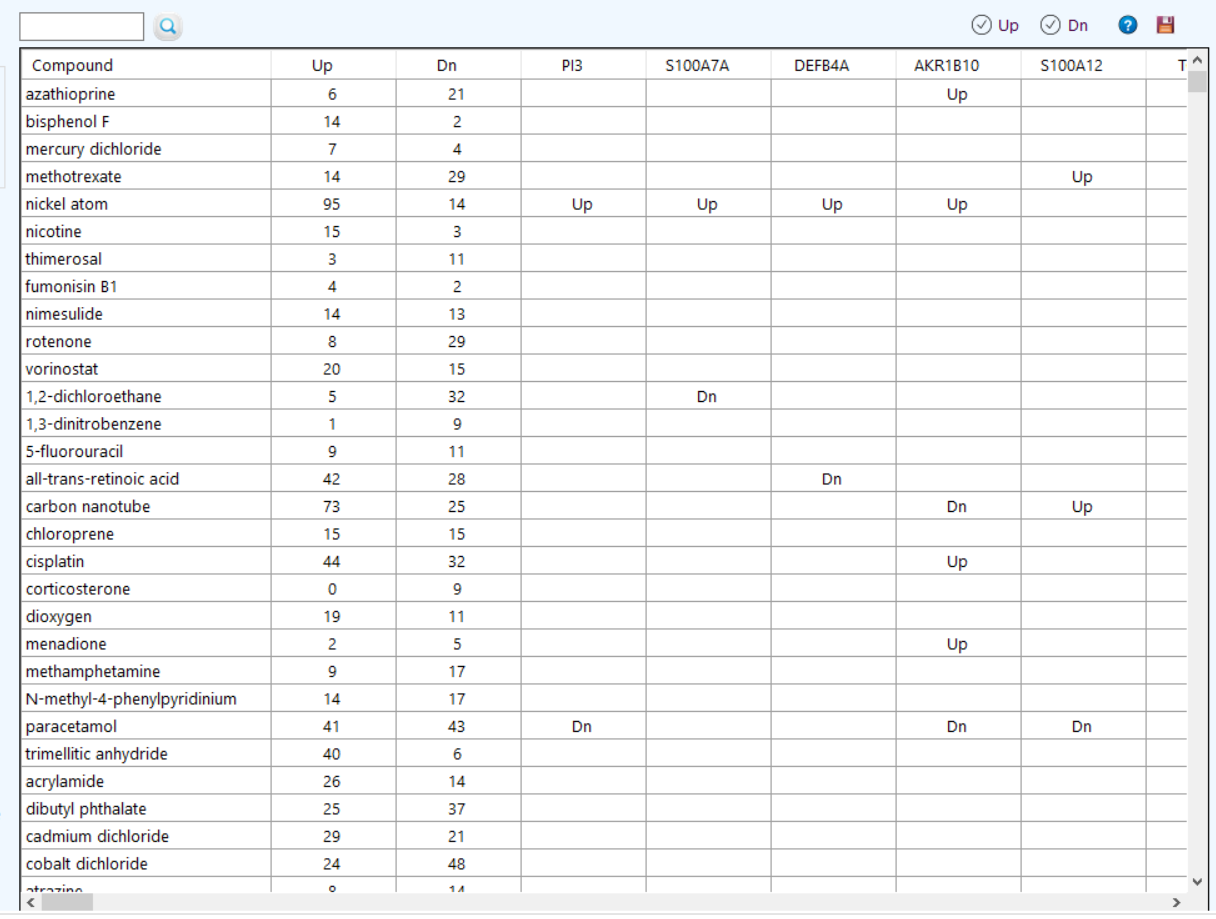
 to see the ChEBI qualifiers.
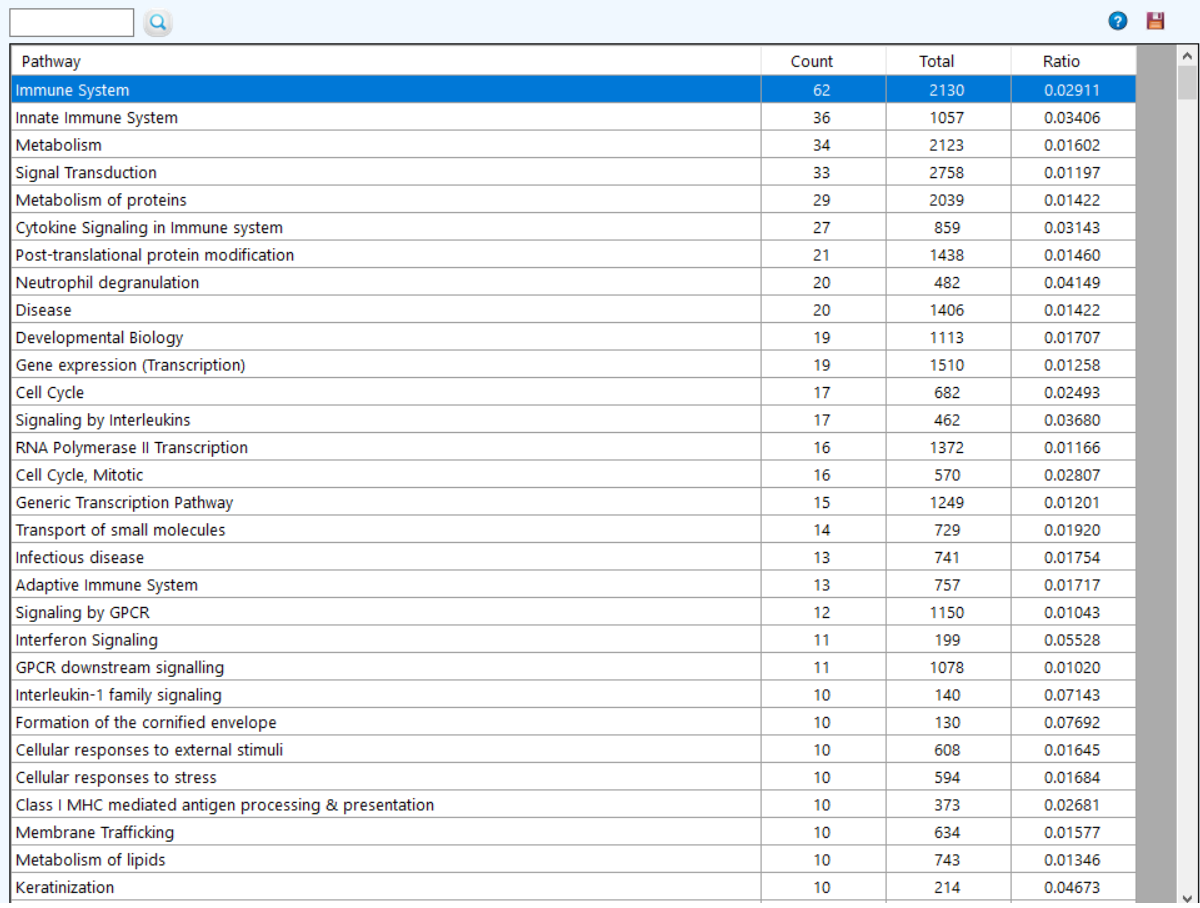
to see the ChEBI qualifiers.  to see the KEGG pathway enrichment.
to see the KEGG pathway enrichment.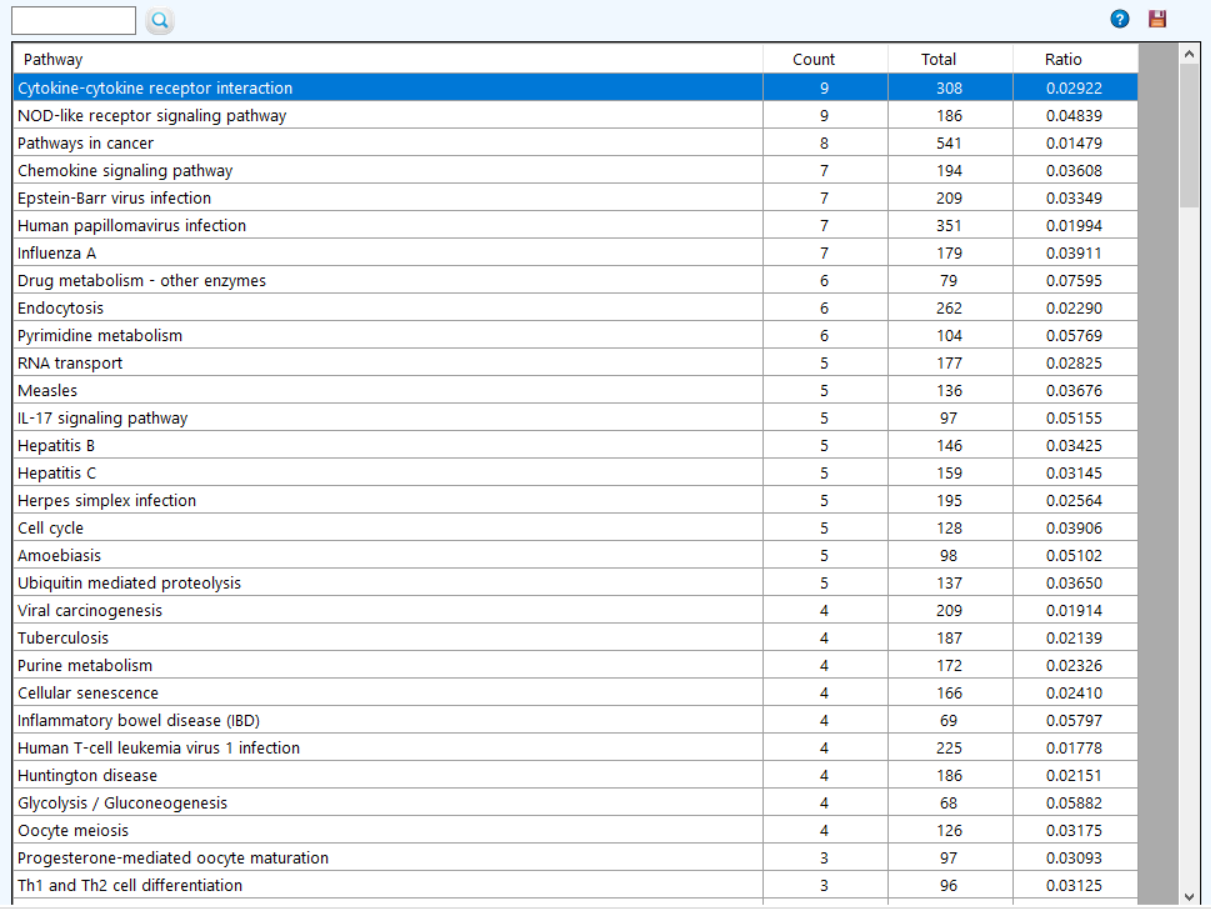
 to see the gene ontology pathway enrichment.
to see the gene ontology pathway enrichment.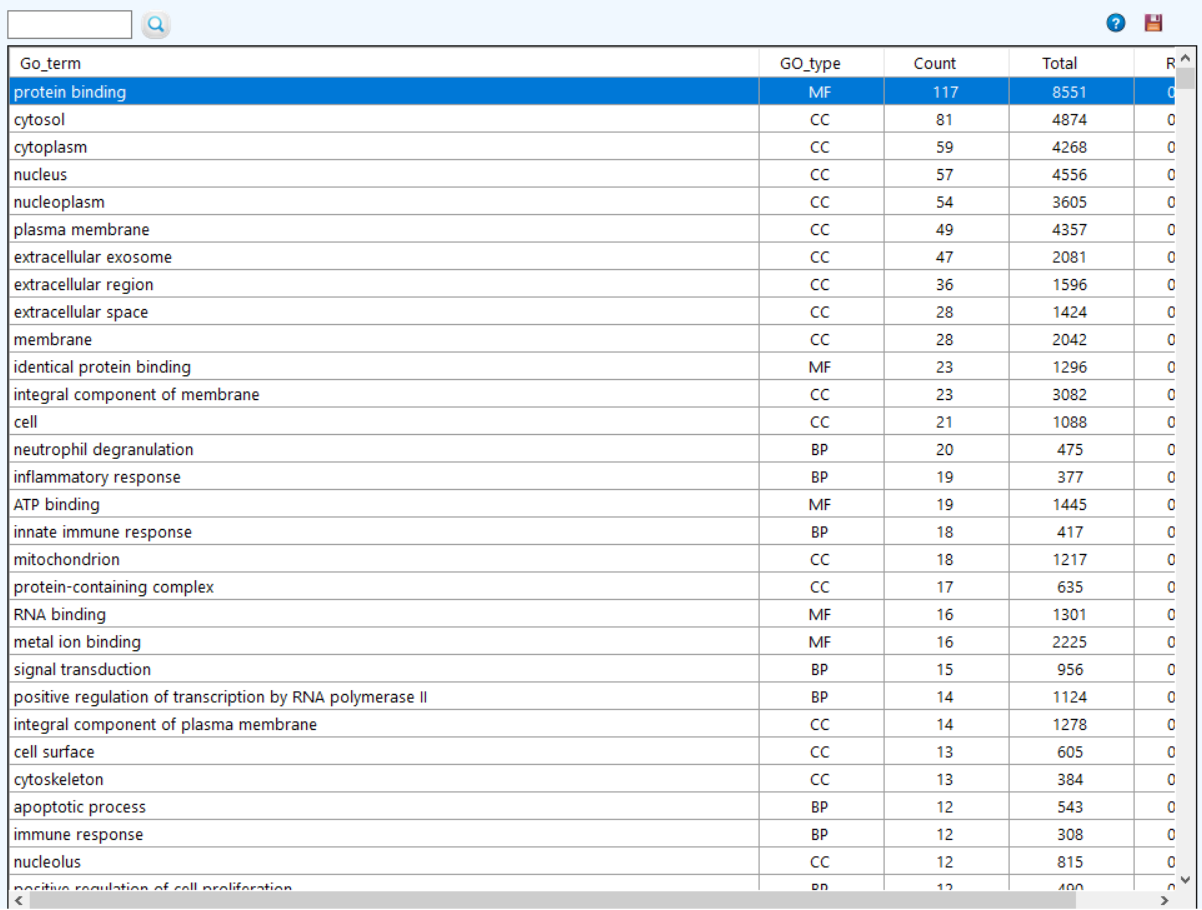
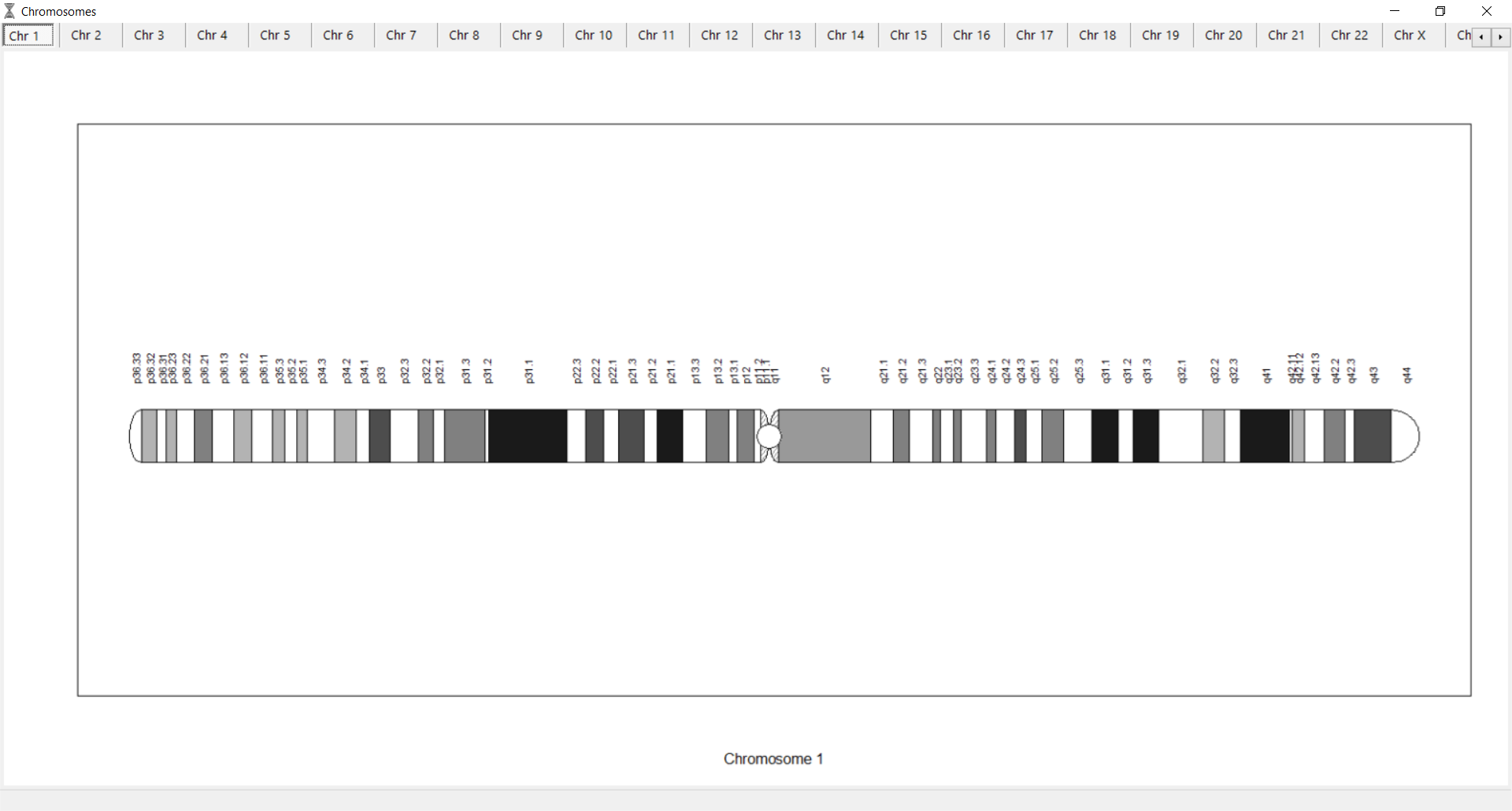
 to see the chromosomes frequency.
to see the chromosomes frequency.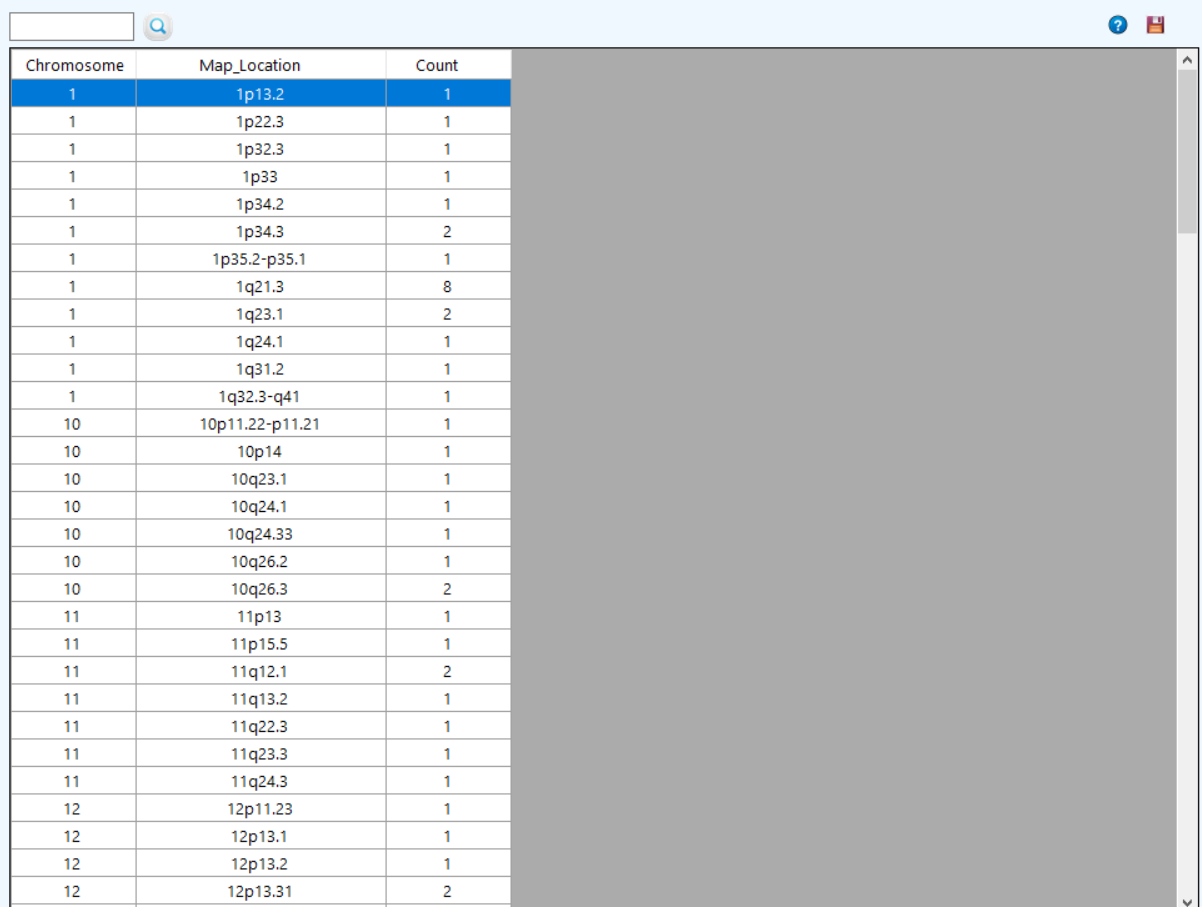
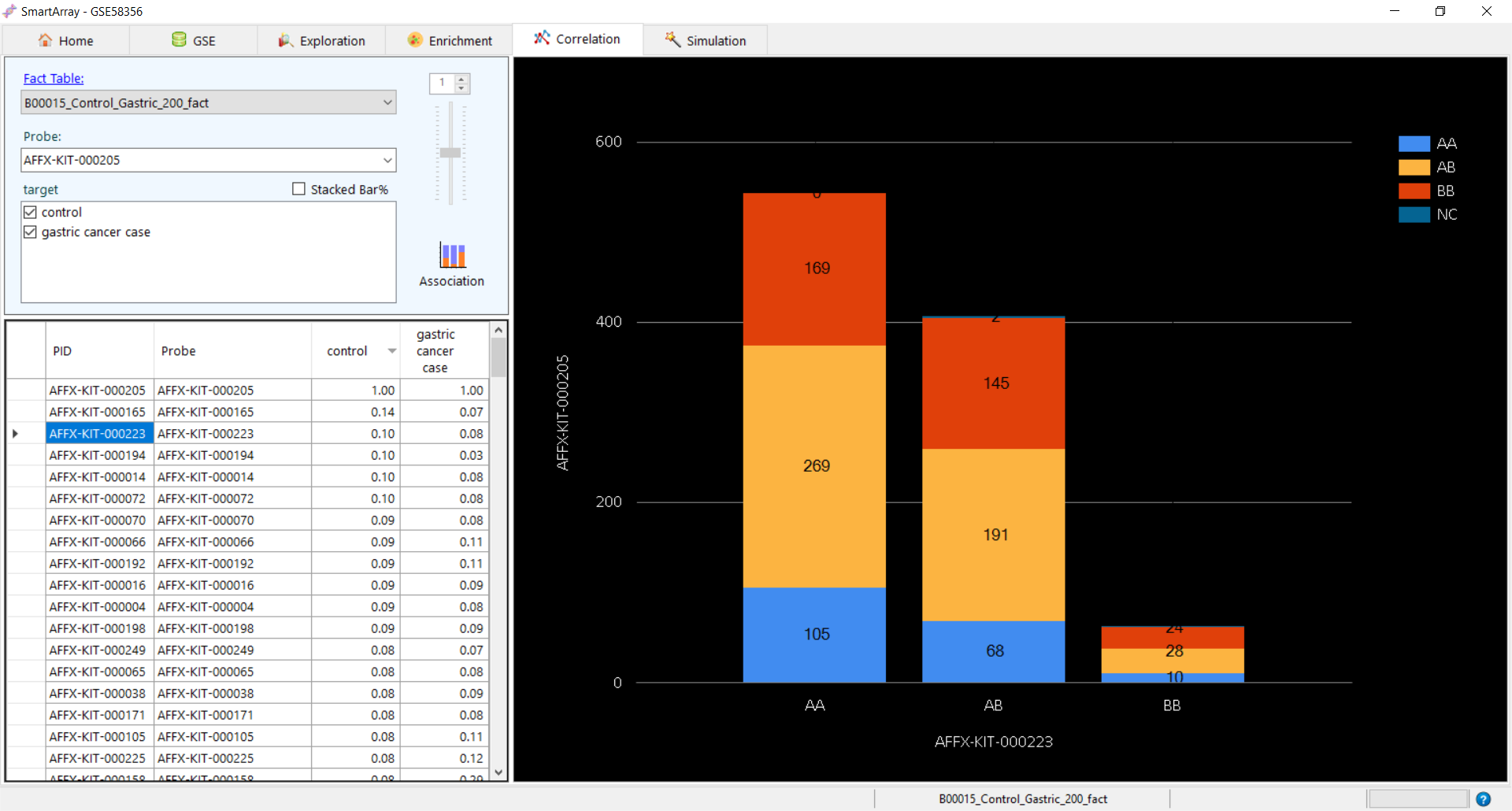
 Association in case of SNPs
Association in case of SNPs 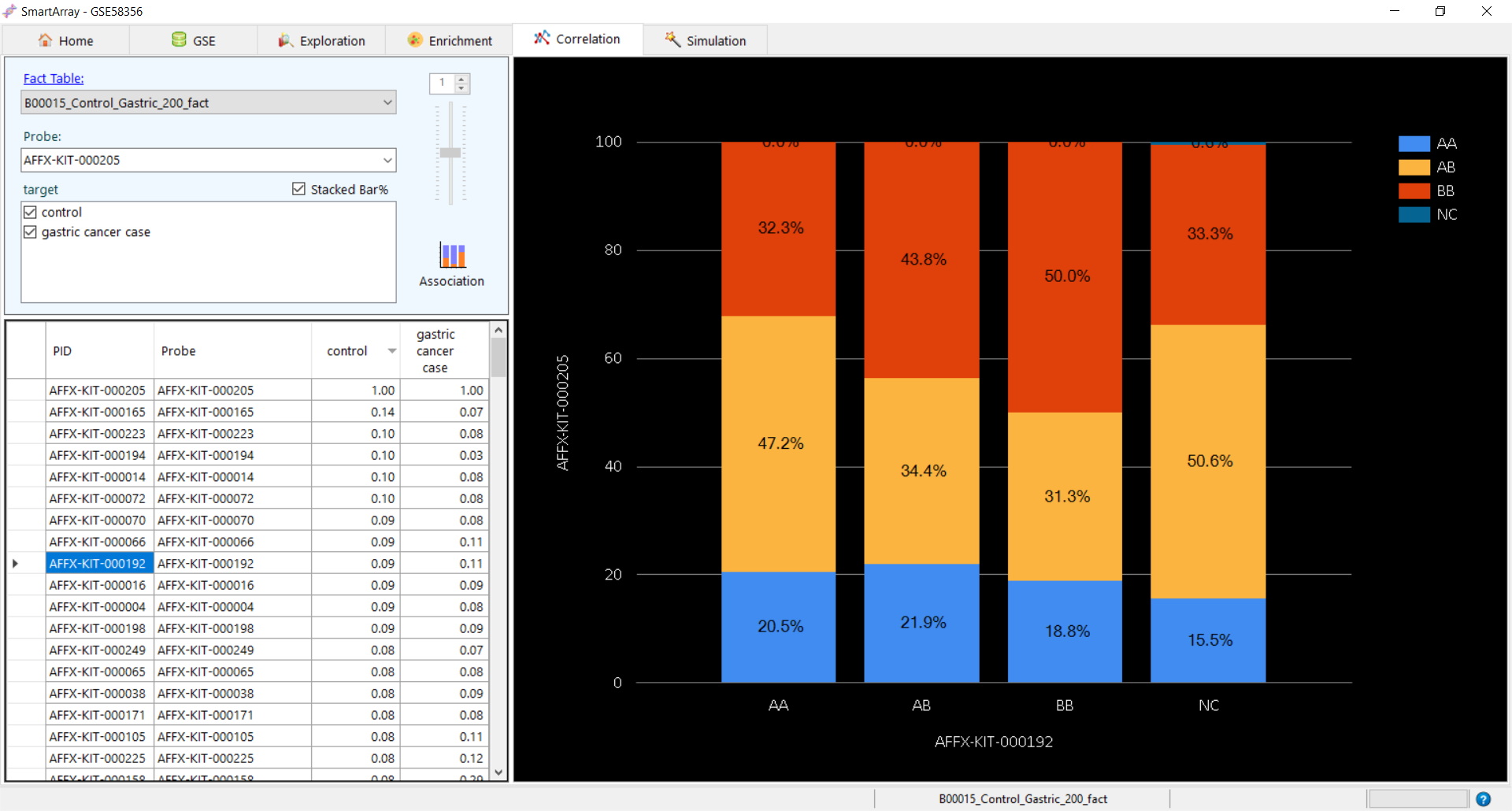
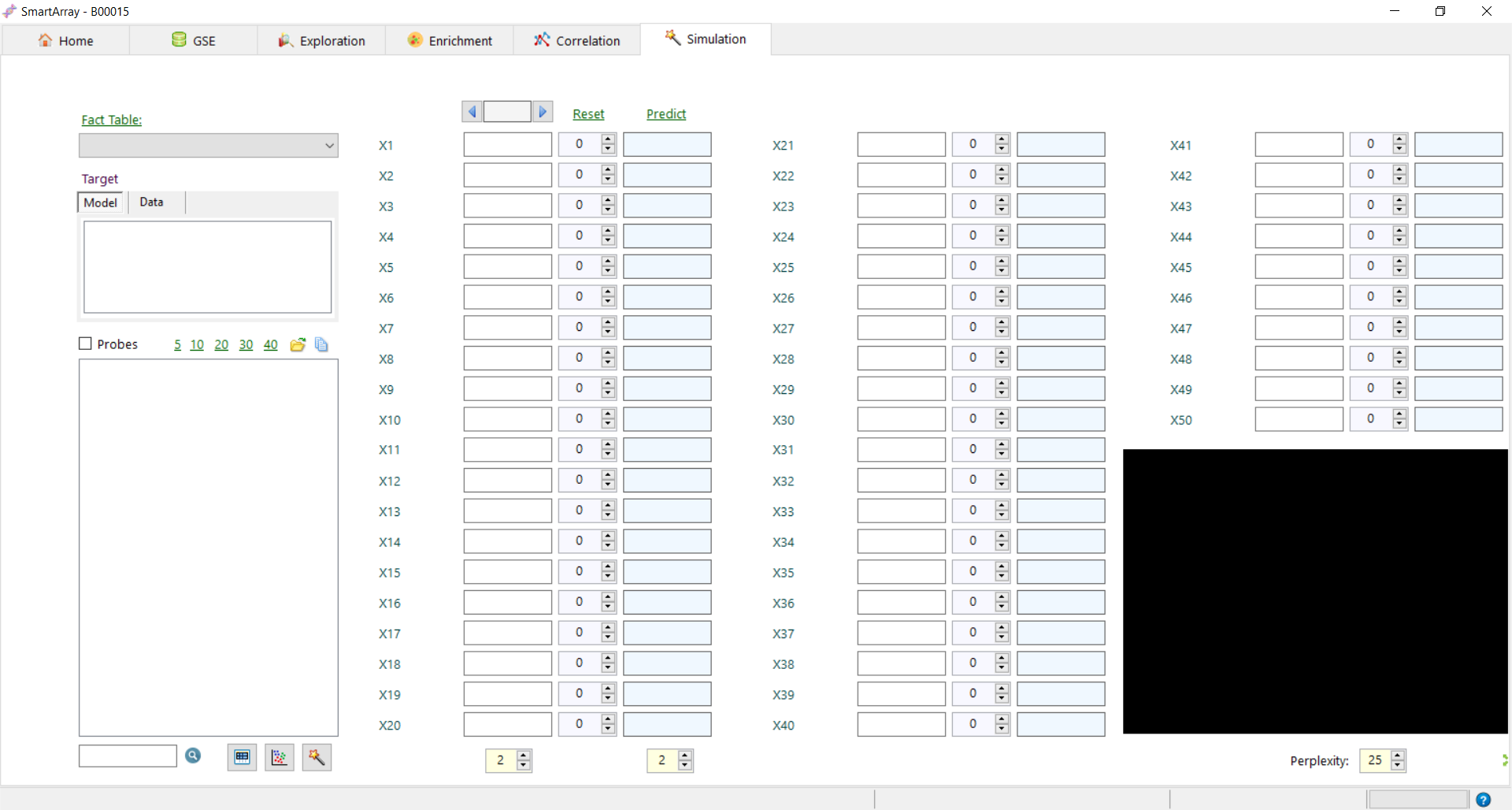
 to build simulation models.
to build simulation models.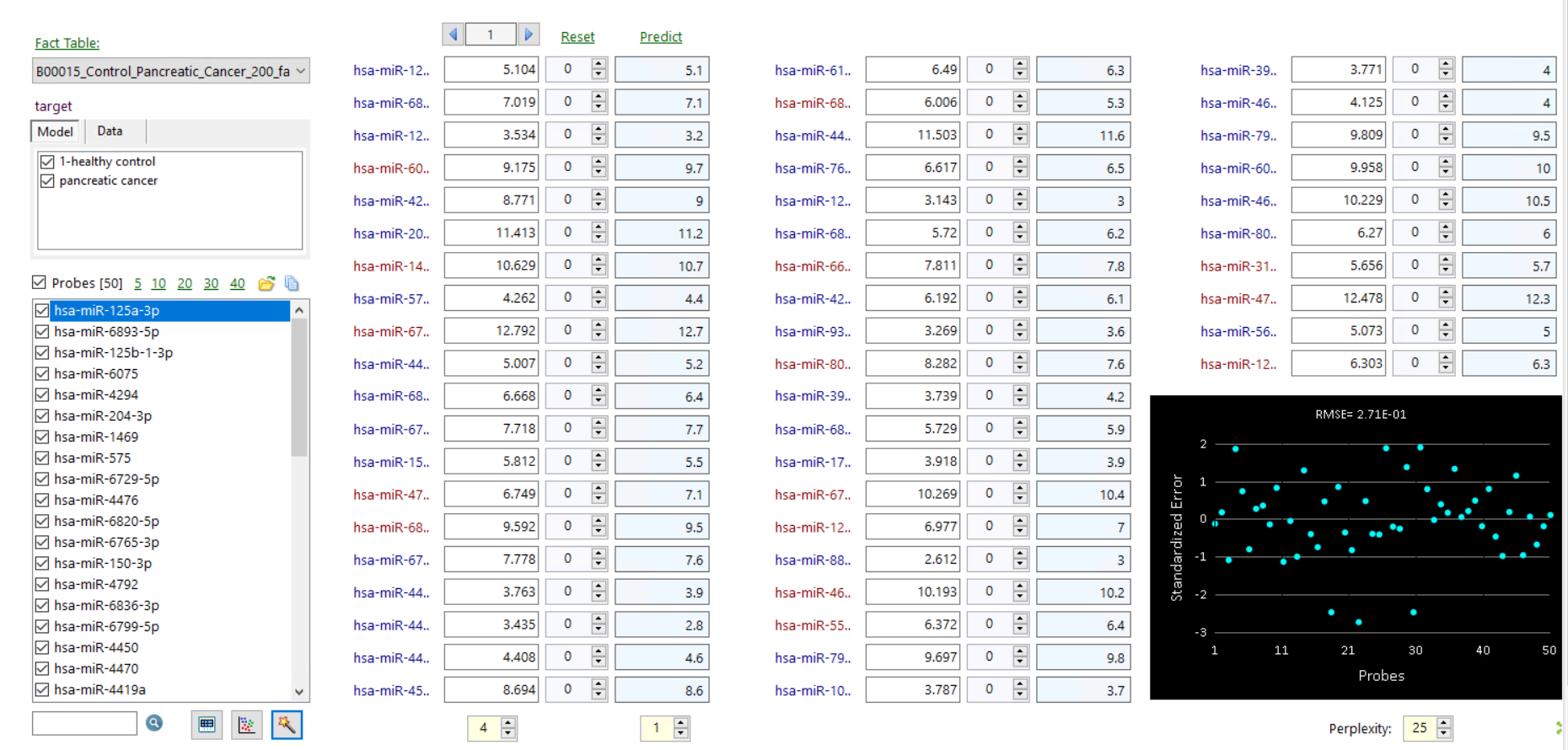
 to display the t-SNE plot.
to display the t-SNE plot.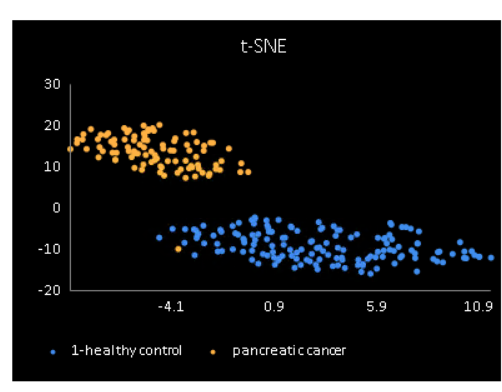
 to show the plots in full screen.
to show the plots in full screen. to get the dataset used for building simulation models.
to get the dataset used for building simulation models.